Intro to Data Science
Lab 3 – Data Wrangling II
A Guide to Your Process
Scheduling
Learning Objectives
Practice
Supporting Information
Class Discussion
Today’s Plan
- Muddiest Point Review
- Intro to the Pipe
- Groupwise Summarization with
dplyr - Discuss Function Tutorial Assignment
Today’s Learning Objectives
After today’s session you will be able to:
- Use the pipe operator in your code
- Perform group summarization with
dplyrfunctions
Muddiest Point Review
- Recurring topics from most recent MPs:
- What other topic(s) would you like to review?
Pipe Operator (%>%)
- Allows chaining together multiple operations
- Product of each function passed to next function
- Same workflow requires fewer objects
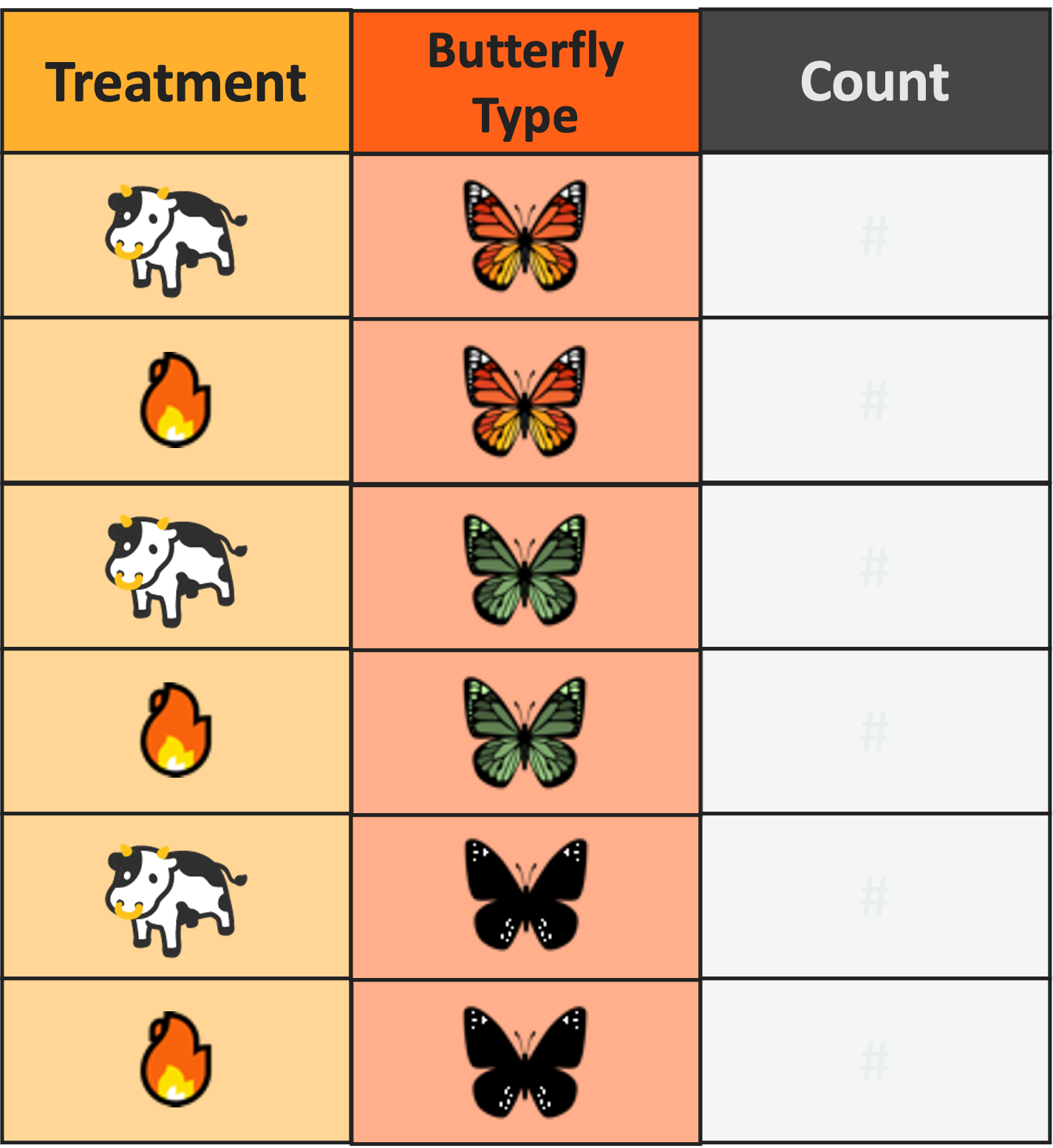
Pipe Operator Example
Without Pipe
# Load data
df_v1 <- read.csv("butterfly.csv")
# Subset to only one treatment
df_v2 <- filter(df_v1, treatment == "cows")
# Add together caterpillars and adult butterflies
df_v3 <- mutate(df_v2, monarch.tot = monarch.bfly + monarch.larva)
# Keep only the total monarch column
df_v4 <- select(df_v3, monarch.tot)Pipe Operator Example
Without Pipe
# Load data
df_v1 <- read.csv("butterfly.csv")
# Subset to only one treatment
df_v2 <- filter(df_v1, treatment == "cows")
# Add together caterpillars and adult butterflies
df_v3 <- mutate(df_v2, monarch.tot = monarch.bfly + monarch.larva)
# Keep only the total monarch column
df_v4 <- select(df_v3, monarch.tot)With Pipe
# Load data
df_v1 <- read.csv("butterfly.csv")
# Do all needed wrangling
df_done <- df_v1 %>%
# Subset to only one treatment
filter(treatment == "cows") %>%
# Add together caterpillars and adult butterflies
mutate(monarch.tot = monarch.bfly + monarch.larva) %>%
# Keep only the total monarch column
select(monarch.tot)Why Named “Pipe”?
René Magritte – The Treachery of Images (1929)



Practice: Pipe

- Install and load the
magrittrpackage
- Return to your 3-step wrangling of “minnow.csv” from Lecture #3
- Filter “minnow.csv” to only Stonerollers and Chubs
- Convert depth & diameter to meters (from cm)
- Pare down columns to only species and depth/diameter in meters
- Copy these lines and edit them to use the
%>%- Does this have the same end result as the non-pipe lines?
Groupwise Summarization
- Summarizing within groups is a common operation
- Average barnacle number at several tidal heights
- Variation in reported customer satisfaction within demographic groups
dplyroffers three functions to accomplish thisgroup_bysummarizeungroup
Summarization Syntax
group_byhas similar structure toselect- Wants column names separated by commas
summarizehas similar structure tomutate- E.g.,
new_column = function(old_column)
- E.g.,
ungrouphas no arguments!
Relevant Helper Functions
- To summarize you’ll need to use functions that calculate summary values
- Take an average with
mean- Has
na.rmargument that determines whether missing values are included
- Has
- Find standard deviation with
sd- Common measurement to use as error bars in a graph
- Find the smallest or largest number with
minandmax
Summarization + Pipes
Let’s check out an example:
Summarization Warnings
- Summarizing simplifies dataframes!
- After summarizing, you’ll have one row per combination of grouping columns
- Summarizing drops columns unless:
- Column is named in
group_by - Column is created by
summarize
- Column is named in
- If you don’t want to lose a column, it needs to meet one of those criteria
Practice: Summarizing



- Using the “penguins” data in the
palmerpenguinspackage, answer the following questions:
- What is the average bill length in millimeters for each species of penguin?
- Which island has the smallest individual penguin?
- Hint: use body mass
- Which species at which island has the longest flippers for female penguins?
- Hint: remember you can use
filterbefore or aftersummarize!
- Hint: remember you can use
Temperature Check
How are you Feeling?
Function Tutorial: Learning Objectives
After completing this assignment you will be able to:
- Explain the proper syntax and use of R functions
- Communicate effectively to an audience of interested non-specialists
- Apply feedback on an assignment to a successful revision
- Reflect on the process of revising a presentation based on constructively critical feedback
Function Tutorial: FAQ
- Tutorial should be Quarto document with plain text and code chunks
- Write tutorials for your classmates for three functions from packages on CRAN
- You’ll present your tutorials for 5-10 minutes in Lab #5
- Get peer feedback then & implement changes before submitting draft 2
- Submit & present revised tutorials during Lab #7
Function Tutorial: Points
- Draft 1 = 30 pts (12% course grade)
- Overall report – 6 pts
- Function tutorial (x3) – 8 pts each
- Draft 2 = 40 pts (16% of grade)
- Overall report – 6 pts
- Function tutorial (x3) – 8 pts each
- Revision response – 3 pts
- Edited from draft 1 from peer feedback – 7 pts
- Optional Draft 3 = 40 pts
- If submitted, score replaces draft 2
- Score can only improve (no way draft 3 reduces total points earned)
Picking Functions
- Everyone must pick three different functions
- This way no two people present tutorials on the same function
- Unfortunately, means if someone picks before you they “claim” that function
- My plan to do this equitably is as follows:
- Randomize student order and each person picks one function
- Second function picked in reverse of that order (I.e., if you were last to pick in round 1, you’re first in round 2)
- Re-randomize student order for third function
- Sound fair? If not, what’s a good alternative?
Nick’s Recommended Packages



stringr – Simple, Consistent Wrappers for Common String Operations
dndR – Dungeons & Dragons Functions for Players and Dungeon Masters
lterpalettefinder – Extract Color Palettes from Photos and Pick Official LTER Palettes


supportR – Support Functions for Wrangling and Visualization
vegan – Community Ecology Package
Forbidden Packages (Sorry!)



dplyr– A Grammar of Data Manipulation- Reason: we cover a lot of this in class
tidyr– Tidy Messy Data- Actually only 2 forbidden functions:
pivot_longer&pivot_wider - Others are okay to use!
- Reason: we just covered both in class
- Actually only 2 forbidden functions:
ggplot2– Create Elegant Data Visualizations Using the Grammar of Graphics- Reason: we cover a lot of this in class (see week 6) and its functions use a really different syntax from what is used by other packages
Assignment Q & A
- What questions do you have about this assignment?
- No such thing as a “dumb” question, so ask away!
- Feeling good about next steps?
Exploring CRAN Packages
- Visit cran.r-project.org
- Click “Packages” on the left sidebar
- Approx. 2/3 down sidebar items
- Click “Table of available packages, sorted by name”
- Scroll through and look for one with a cool name / title!
Practice: Exploring CRAN
- Explore available packages / functions
- Select 7-10 functions so you have alternates (if needed)
- We will pick functions during next lecture (Lecture #4)
Temperature Check
How are you Feeling?

Upcoming Due Dates
Due before lecture
(By midnight)
- Homework #3
- Pick 7-10 possible functions for Function Tutorial assignment
- Remember, they must be from CRAN packages!
Due before lab
(By midnight)
- Muddiest Point #4
Back to Course Page: Programming in R for Biologists
Bonus: Data Shape
Bonus Learning Objectives
After this bonus session you will be able to:
- Explain what “shape” means in the context of data
- Reshape data from long to wide format
- And vice versa
Data “Shape”
- Data with rows/columns has a shape
- Shape refers to whether observations are in the rows or the columns
- “Wide” data has observations as columns
- E.g., Each column is a different species’ count
- “Long” data has observations as rows
- E.g., The columns are “species” and “count”
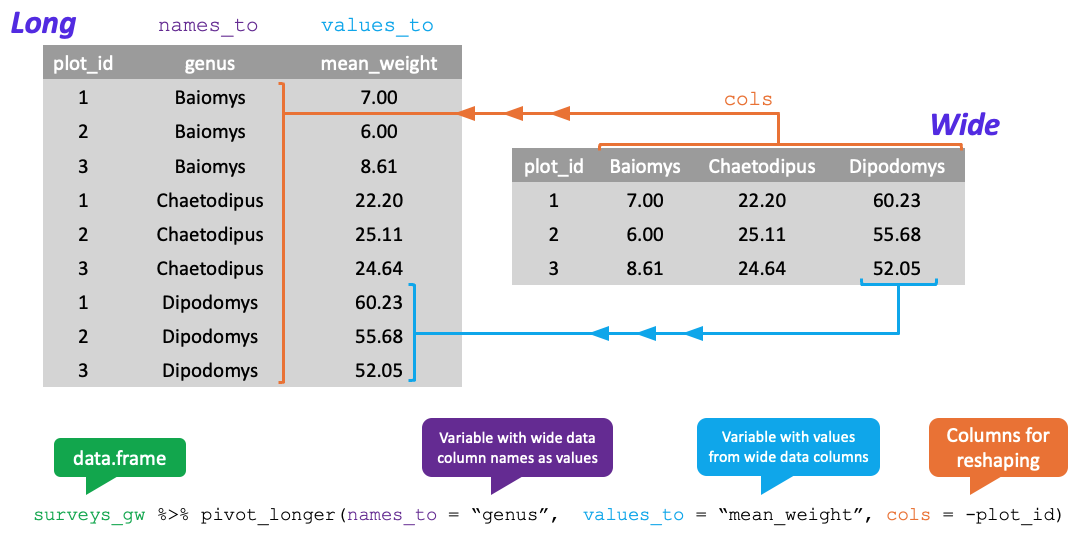
Data Shape Visual
Long Data
Wide Data

Reshaping Longer
- Change from wide to long format with
tidyr::pivot_longer- Has 4 key arguments
data= the wide data to pivot
cols= the columns to pivot- Can select which columns to pivot OR which to not
- Include:
cols = colD:colX - Exclude:
cols = -colA:-colC
Reshaping Longer Continued
names_to= name of new column to hold old column names- Must be in quotes
values_to= name of new column to hold values- Also in quotes
Reshaping Longer Visual
Practice: pivot_longer

- Download the “bees.csv” and load it into R with
read.csv- Check its structure! What columns are there?
- Pivot the data so that you are left with three columns:
- “year”, “bee_group”, and “bee_abundance”
- Check your work! What are the dimensions of the resulting dataframe?
- Should be 32 rows by 3 columns
Reshaping Wider
- Change from long to wide format with
tidyr::pivot_wider- Also has 4 key arguments
data= the wide data to pivot
names_from= name of the column to turn into new column names- Must be unquoted
Reshaping Wider Continued
values_from= name of column to make into new column values- Also unquoted
values_fill= value to fill if value is missing in original data- Technically optional but good practice to include explicitly
Reshaping Wider Visual
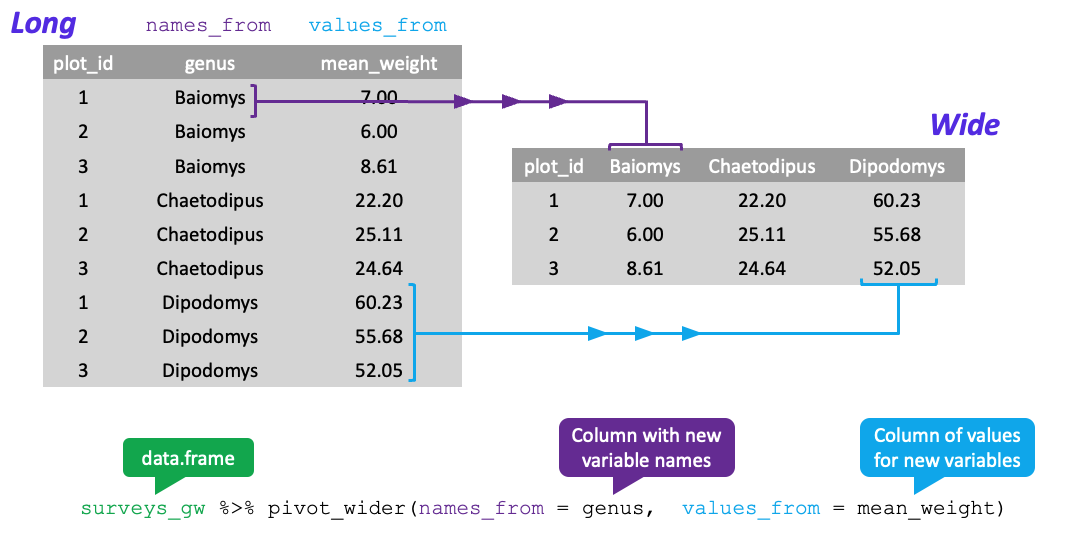
Practice: pivot_wider

- Take the data object you pivoted to long format in the prior practice block
- Pivot it back to wide format with
pivot_wider!
- Check your work!
- Does it look like the original object you loaded with
read.csv?
- Does it look like the original object you loaded with
Practice: Wrangling!




- Beginning with the “penguins” data do the following operations:
- Keep only data on female penguins
- No male penguins and no individuals where sex is not known
- Calculate average bill depth within species and island
- Reshape to wide format so that each island is a column
- Note that if an island doesn’t have a given species it should have
NA(not0)
- Note that if an island doesn’t have a given species it should have
- Check your work! What are the dimensions of the resulting dataframe?
- Should be 6 rows by 5 columns