Intro to Data Science
Lecture 3 – Quarto Files & Data Wrangling I
A Guide to Your Process
Scheduling
Learning Objectives
Practice
Supporting Information
Class Discussion
Today’s Plan
- Quarto Files
- Loading Data
- Working with Data
- Tidyverse -
dplyr
Today’s Learning Objectives
After today’s session you will be able to:
- Define the three major components of Quarto files (.qmd)
- Write code to load external data into R
- Explore data with base R tools
- Manipulate data with
dplyr
GitHub Review
- Great work last week!
- In the next week or two (i.e., before the second half of the course) I would like you to:
- Make another practice repository (or maybe 2!)
- What questions do you have about this?
- Does this feel reasonable to you?
Quarto Files Intro
Quarto (qmd) files have three sections:
- Metadata (YAML)
- Controls formatting of document
- Plain Text
- Technically written in markdown (a text-formatting language)
- Code chunks
- Essentially mini R scripts within the larger file!
Quarto Analogy




Quarto Part 1: Metadata
- Document formatting metadata is called YAML
- Yet Another Markup Language
- Defines document header information & formatting
- Title, Author, Date
- File output type
- Output options:
- HTML = like a webpage but outputs as a file rather than a living website
Quarto Part 2: Plain Text
- Write text just like you would in MS Word / etc.
- But, there is no toolbar with buttons for doing formatting
- Instead markdown syntax is required to accomplish these tweaks
Markdown Syntax
- Your function tutorials have four required markdown styles:
# = headings
- More # = smaller heading
_text_ = italics
**text** = bold
[text](link) = hyperlinked text
Other format options here: markdownguide.org/basic-syntax
Quarto Part 3: Code Chunks
Let’s look at the structure of an example code chunk
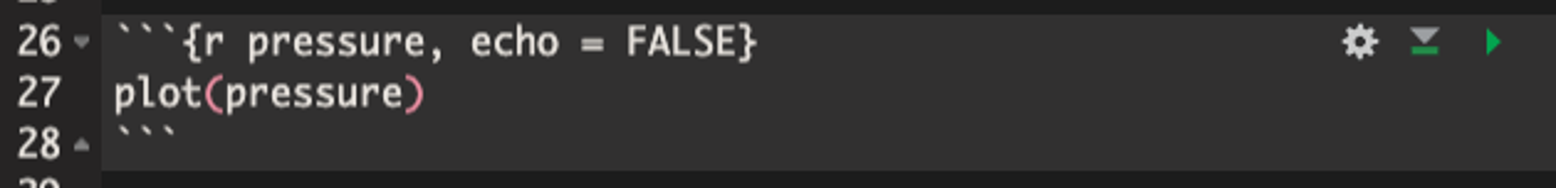
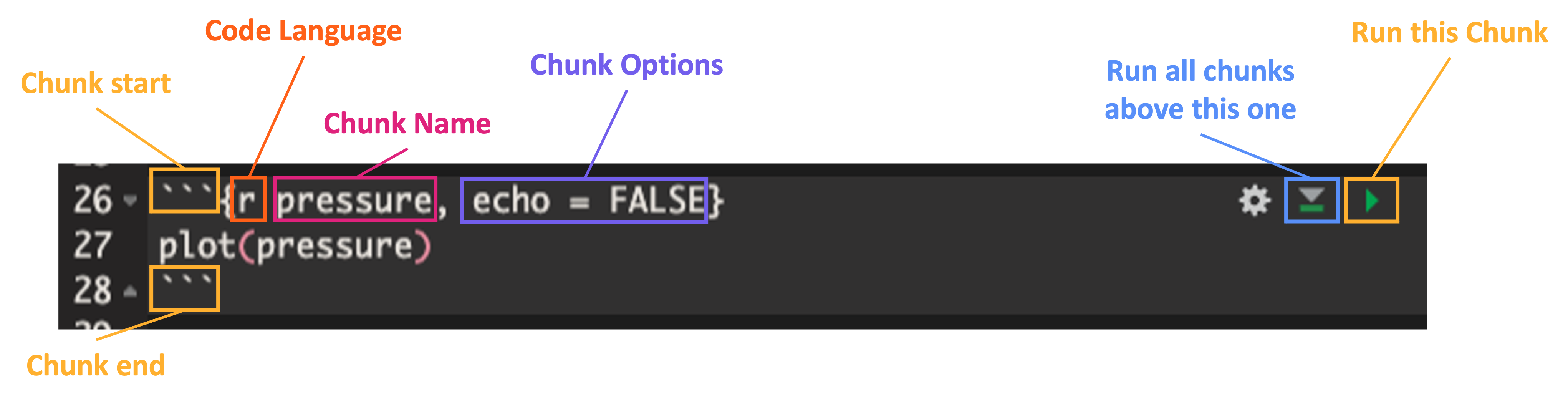
Note that chunk start must be formatted like:
- ```{language chunk_name, option_1, option_2, ...}Code Chunks Options
Let’s check out three crucial code chunk options!
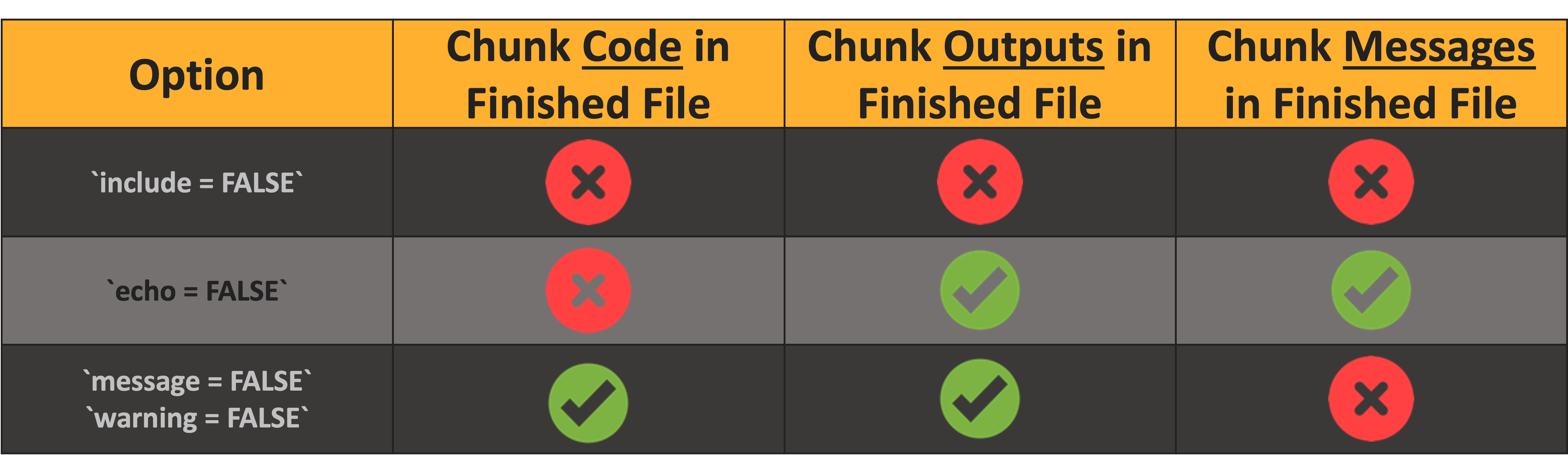
- For a full list of options see here
Practice: Quarto Files

- Create a new Quarto file!
- File New File Quarto Document…
- In resulting pop-up, skip to bottom and click “OK”
Look at (1) YAML, (2) Plain text, and (3) code chunks
- Take notes on anything that jumps out at you
- Click the “render” button
Temperature Check
How are you Feeling?
Loading Data
- Function to use depends on file type
- CSV =
read.csv - MS Excel =
readxl::read_excel
- CSV =
- Need to assign data to an object to use it later!
Download Example Data
- From the course page, download “minnow.csv”
- Move “minnow.csv” from your “Downloads” folder to your RStudio Project folder
- Make a new script for today’s lecture
Practice: Load Data

- Now, use
read.csvto read “minnow.csv” into R- Remember to assign it to an object!
- First thing after reading in data: check structure!
- Can use
strordplyr::glimpse
- Can use
- What do you see?
Exploring Data with Base R
Two ways in base R to access data:
- Bracket notation (works similar to vectors)
- Dollar sign ($) notation
Bracket Notation
- The syntax is:
data[row number, column number]
- Let’s look at some example cases
- Note that concatenation works here too!
my_df[c(1, 2, 3), 1]would get rows 1 through 3 of column 1
Dollar Sign Notations
- The syntax is:
data$column
- Note that this does not work for rows!
Practice: Base R Data Exploration
- Using bracket notation:
- Access the 7th row of the minnow data
- Access the 5th column of the minnow data
- What is the value in the 21st row and 3rd column?
- Using dollar sign notation:
- Check the “diameter” column
- Look at the “species” column
Tidyverse Background
- Ecosystem of inter-related packages & functions
- Very human-readable
- Extremely popular & commonly-used








dplyr Part 1: filter
- Remember our discussion of conditionals last week?
- Types include:
==,|, and&
- Types include:
- Subset using conditionals with
filterdplyr::filter==subset
- Can use
filterinstead ofsubsetjust to live fully in the Tidyverse- Just a style choice, so your call!
dplyr Part 2: mutate
- Make new columns with
mutate
- Can create multiple columns at the same time
Column Naming Aside
- Avoid spaces or hyphens (
-) in column names- Programming languages don’t like these characters in column names
dplyr Part 3: select
- Pick columns to keep or remove with
select
- Can choose columns to keep or to remove
- Notice that column names are not in quotes
- This is one of the special properties of the Tidyverse
Practice: Wrangling with dplyr

- Filter the minnow data to only cases where the species is Stoneroller or Chub
- For that subset, make new columns where river depth and fish nest diameter are in meters
- Next, keep only the transect, species, diameter in meters, and depth in meters columns
- There are two ways of doing this; can you identify them both?
- Check your work! What are the dimensions of the final data object?
- Should be 14 rows and 4 columns
Temperature Check
How are you Feeling?

Upcoming Due Dates
Due before lab
(By midnight)
- Muddiest Point #3
Due before lecture
(By midnight)
- Homework #3
- Pick 7-10 possible functions for Function Tutorial assignment
- Visit: cran.r-project.org
- Click “Packages” in left sidebar
- Click “Table of available packages, sorted by name”
- Your possible functions must be from these packages!
Back to Course Page: Programming in R for Biologists