# Install librarian (if you need to)
# install.packages("librarian")
# Install (if not already present) and load needed libraries
librarian::shelf(vegan, RRPP, scatterplot3d, TeachingDemos, supportR)Multivariate Statistics 101
Nick J Lyon
Caveat Before We Begin
- Read this book
- Has a complete R appendix for:
- Every example
- Every figure
- Every operation
- Essentially the book is written in R Markdown
- Bonus: actually pretty engaging to read!
- Despite subject matter

Tutorial Outline
Background
Resampling & Permutation
Multivariate Data Visualization
Principle Components Analysis
Non-Metric Multidimensional Scaling
Multivariate Background
- Multivariate data have more variables (p) than observations (q)
- I.e., more columns than rows
- True of most ecology/evolution datasets
- Differs from univariate statistics
- Univariate explores variation in one variable
- Multivariate explores variation in many variables (plus potential inter-relationships)
Resampling Methods
- Frequentist statistics uses distributions from theory
- Resampling statistics uses distributions from data
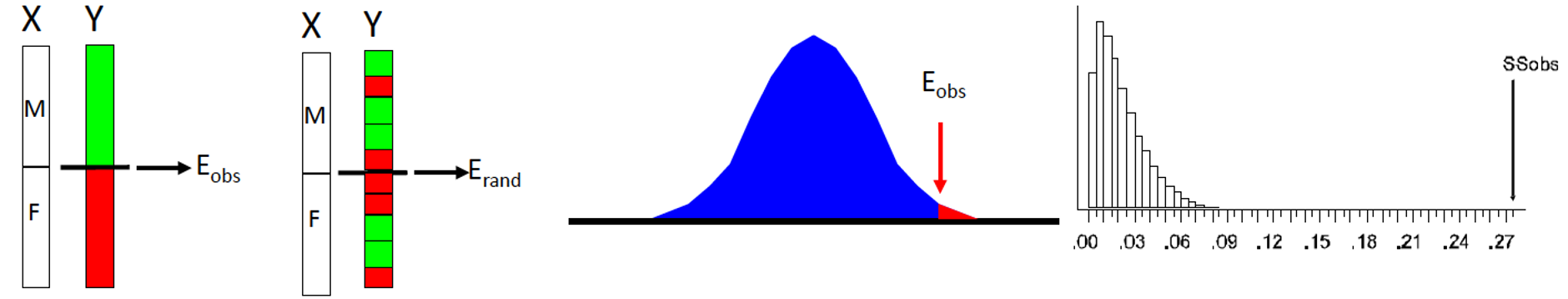
Theoretical Process
- Take samples from data (i.e., “re-sample”)
- Compare real observations to re-sampled groups
- Evaluate significance
Permutation Notes
- Permutation methods are non-parametric
- Because they don’t rely on a theoretical distribution
- Permutation methods are flexible
- Can assess standard & non-standard experimental designs
- Handle high-dimensional data (more variables than observations)
Two Major “Flavors”
Full Permutation
- Permute whole dataset
Residual Permutation
- Fit desired model
- Permute the residuals
- Less sensitive to outliers
Multivariate Visualization
- Typically involves “ordination”
- Frequently uses “multidimensional scaling”
- I.e., getting from many variables to fewer, more easily visualizable variables
- Still representative of multivariate nature of data
- Common ordination methods include:
- Principal Components Analysis (PCA)
- Principal Coordinates Analysis (PCoA)
- Nonmetric Multidimensional Scaling (NMS)
Principle Components Analysis
- Goal: reduce number of variables
- Mechanism: create combinations of existing variables to summarize variation
- Want each combination to contain as much variation as possible
- Such that you approach 100% variation summarized in only a few combinations
- Result: number of principal components equal to number of observations
- Each principle component has a known % variation explained
PCA Process
- For variables (Xi) you want to create indices (Ik)
- Consider the following example:
- I1 = X1 + X2 + X3 + X4 + X5
- I2 = X1 - X2 + X3 + X4 + X5
- I3 = X1 - X2 - X3 + X4 + X5
- …
- Ik = X1 - X2 - X3 - X4 - X5
PCA Special Consideration 1
- Axis orthogonality
- Axes are “constrained to orthogonality” because of goal of maximized explained variation
- Plain language: PC axes are perpendicular to one another
- Means PC3 through PCn are defined as soon as PC1 and PC2 are
- Focusing on early PCs reduces the relevance of this issue
PCA Special Consideration 2
- Not a hypothesis test
- PCA is great for visualizing patterns in data
- Not good for statistical evaluation
- I.e., PCA cannot–by itself–show support for your hypothesis
Nonmetric Multidimensional Scaling
- Goal: reduce number of variables
- Same as PCA!
- Mechanism: scale dissimilarity of points to minimize “stress”
- “dissimilarity” != “distance”
- Stress is a metric for tension between true spatial configuration of points versus the arrangement of their dissimilarity
- Result: number of NMS axes is defined by the user
- NMS reports stress of “best” solution (essentially a goodness of fit metric)
NMS Process
- Choose a starting configuration of points (randomly)
- Move points around and measure stress at each configuration
- Repeat until stress has been minimized
- Return to step 1 with different starting points
- Necessary to avoid local stress minima
- Continue 1-4 until confident true minimum stress configuration has been found
NMS Helicopter Analogy




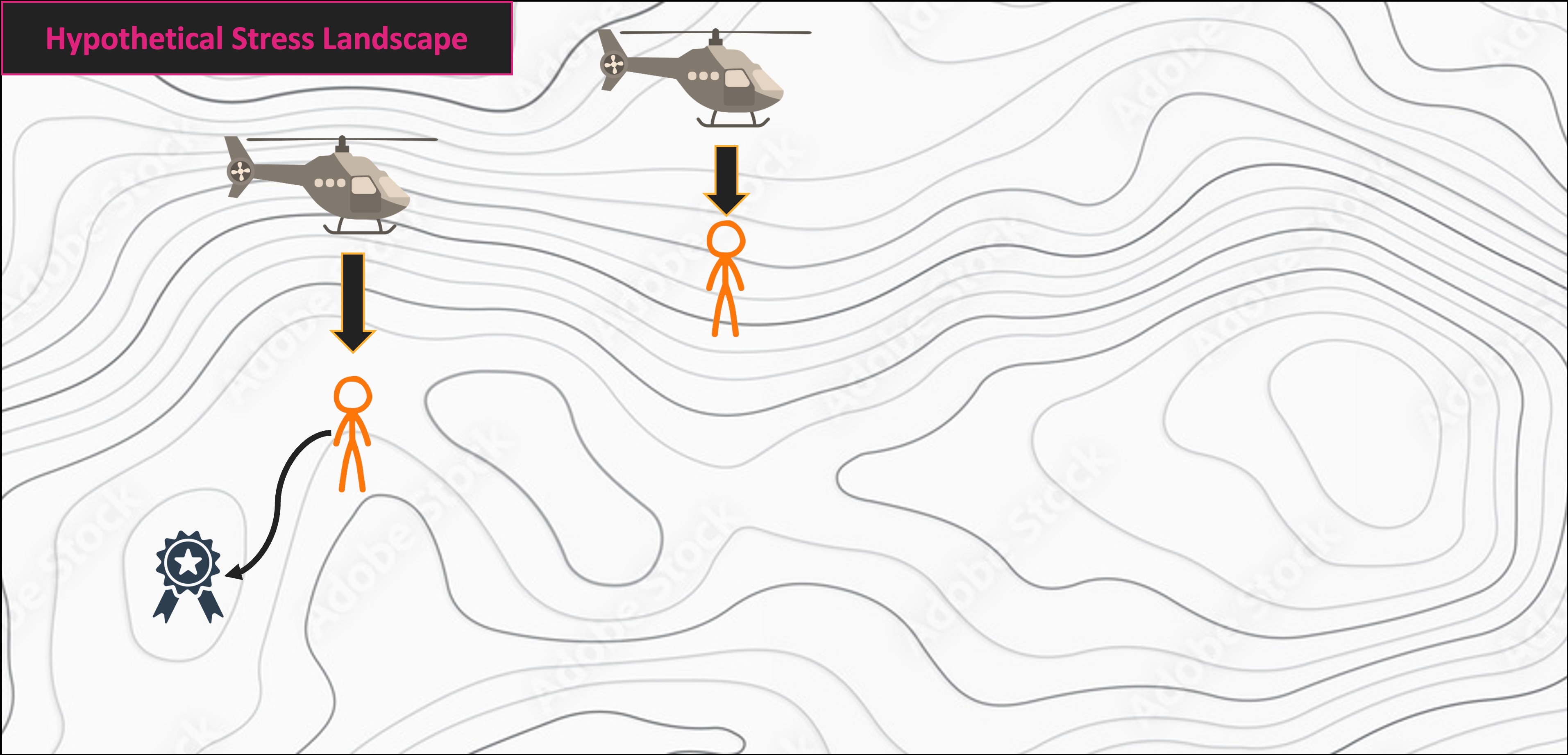
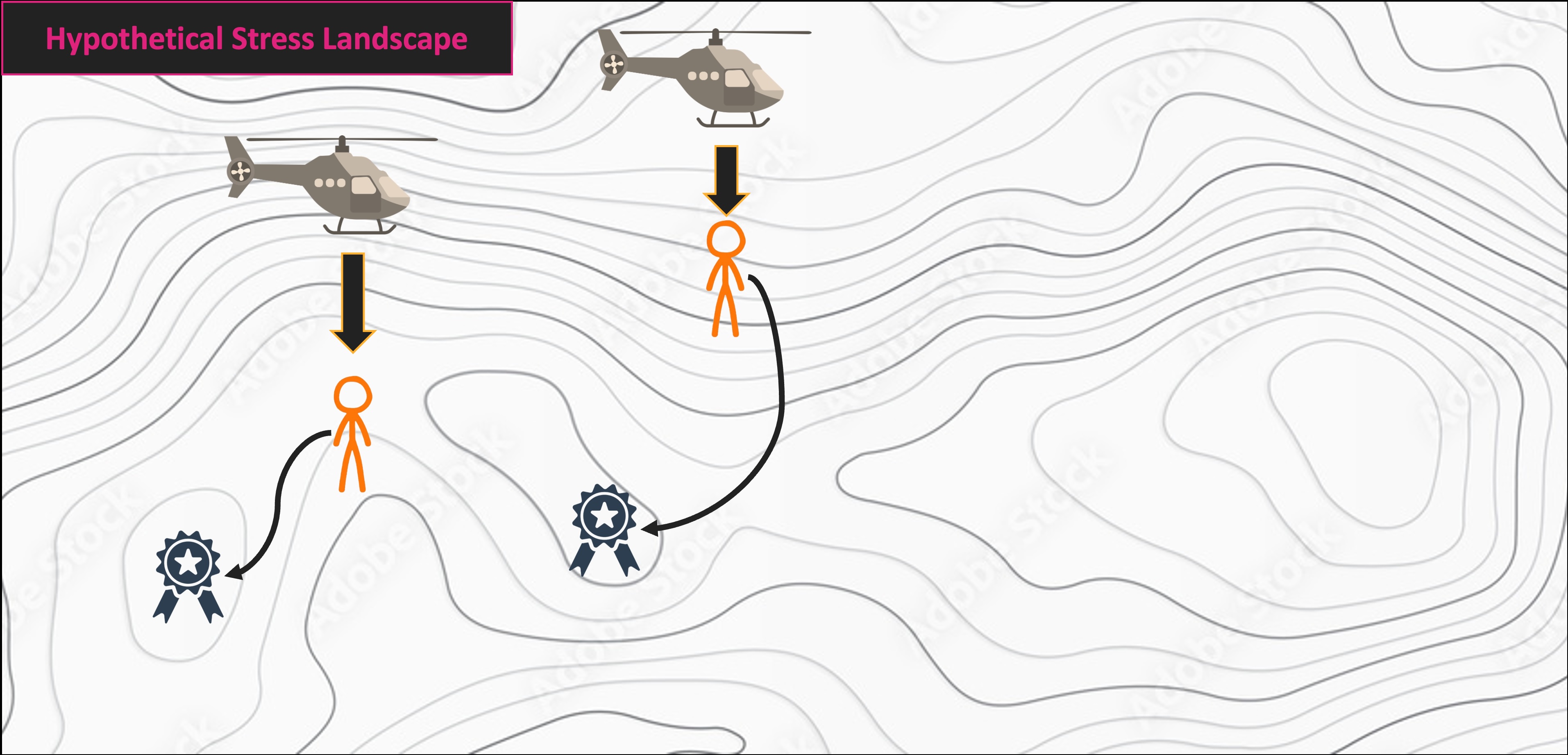
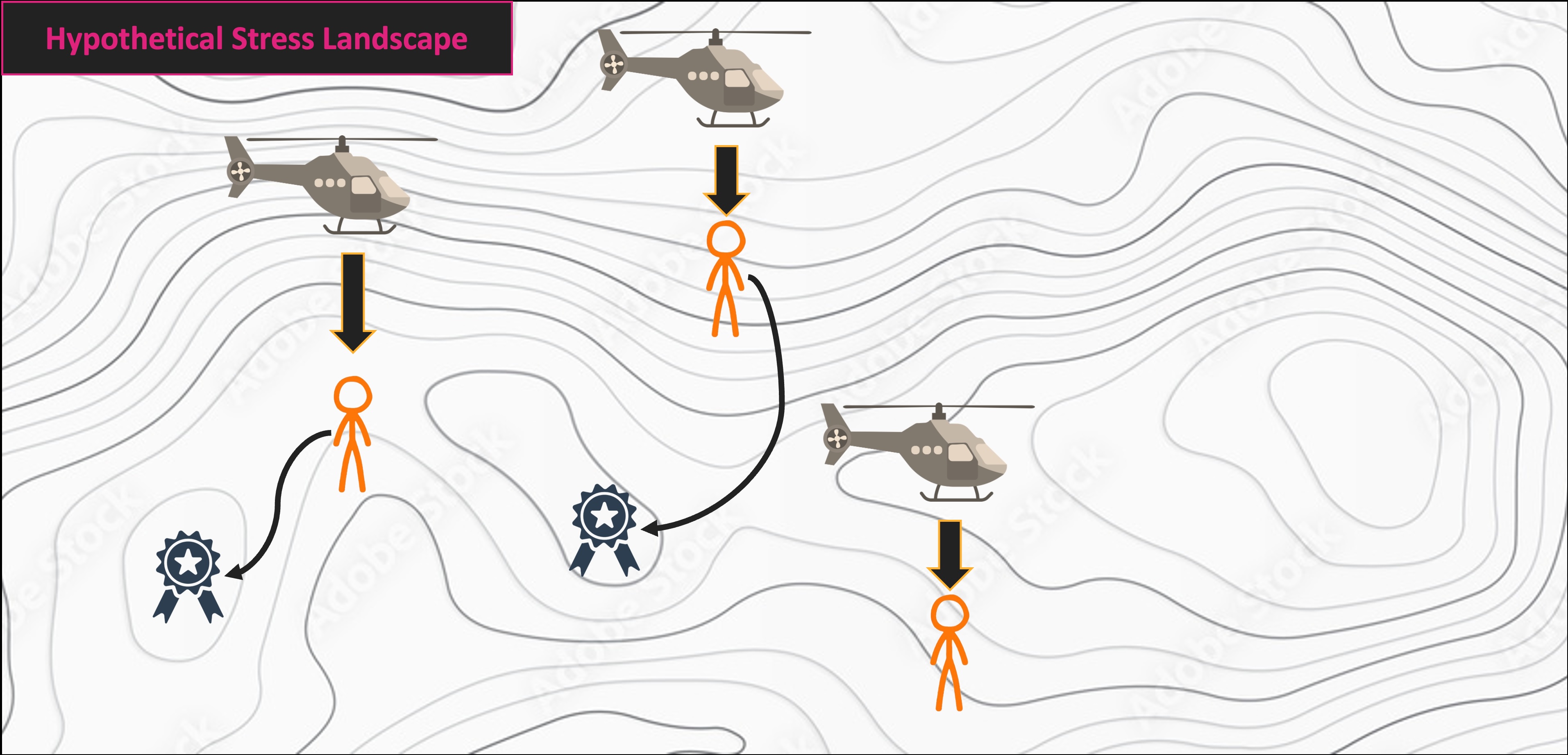
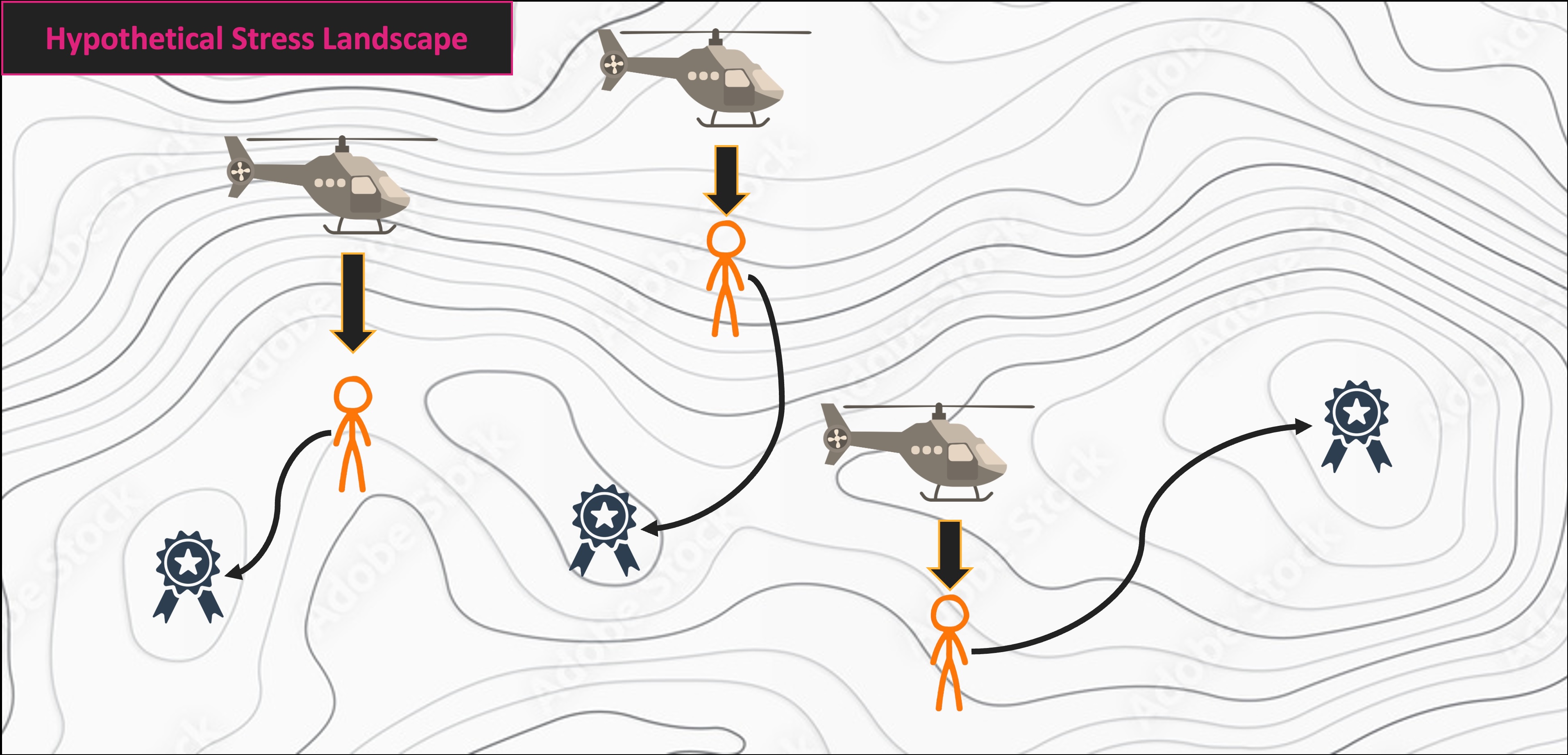
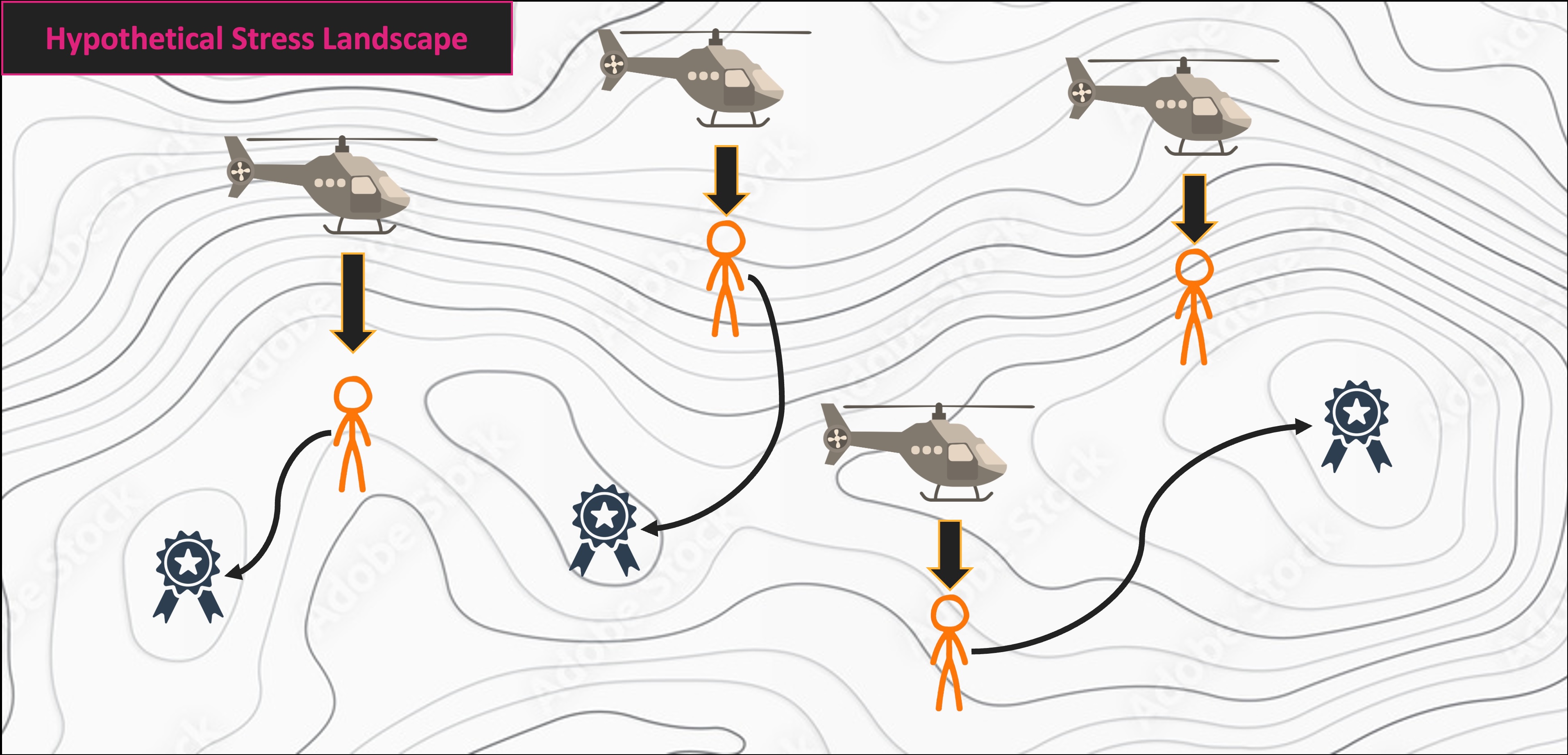
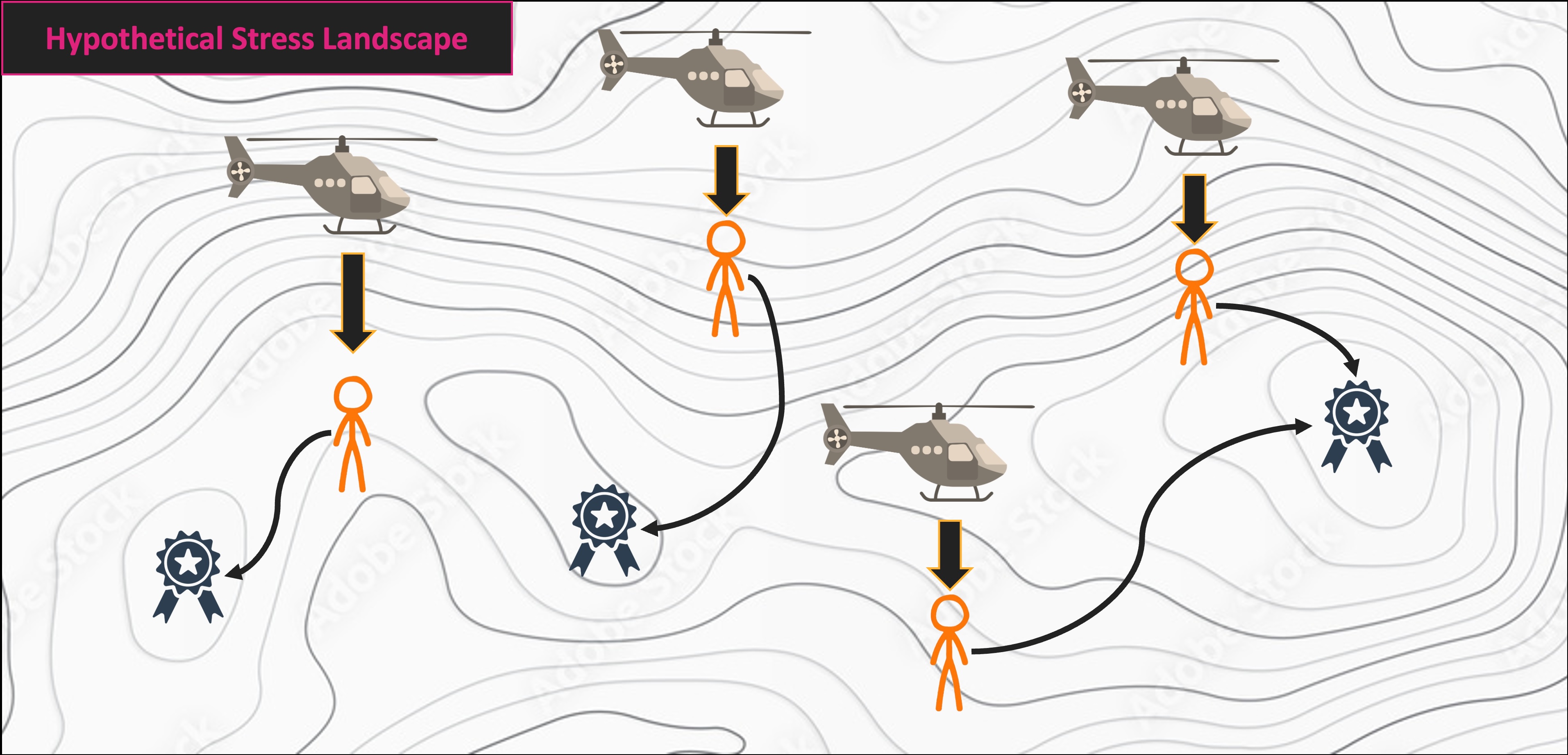
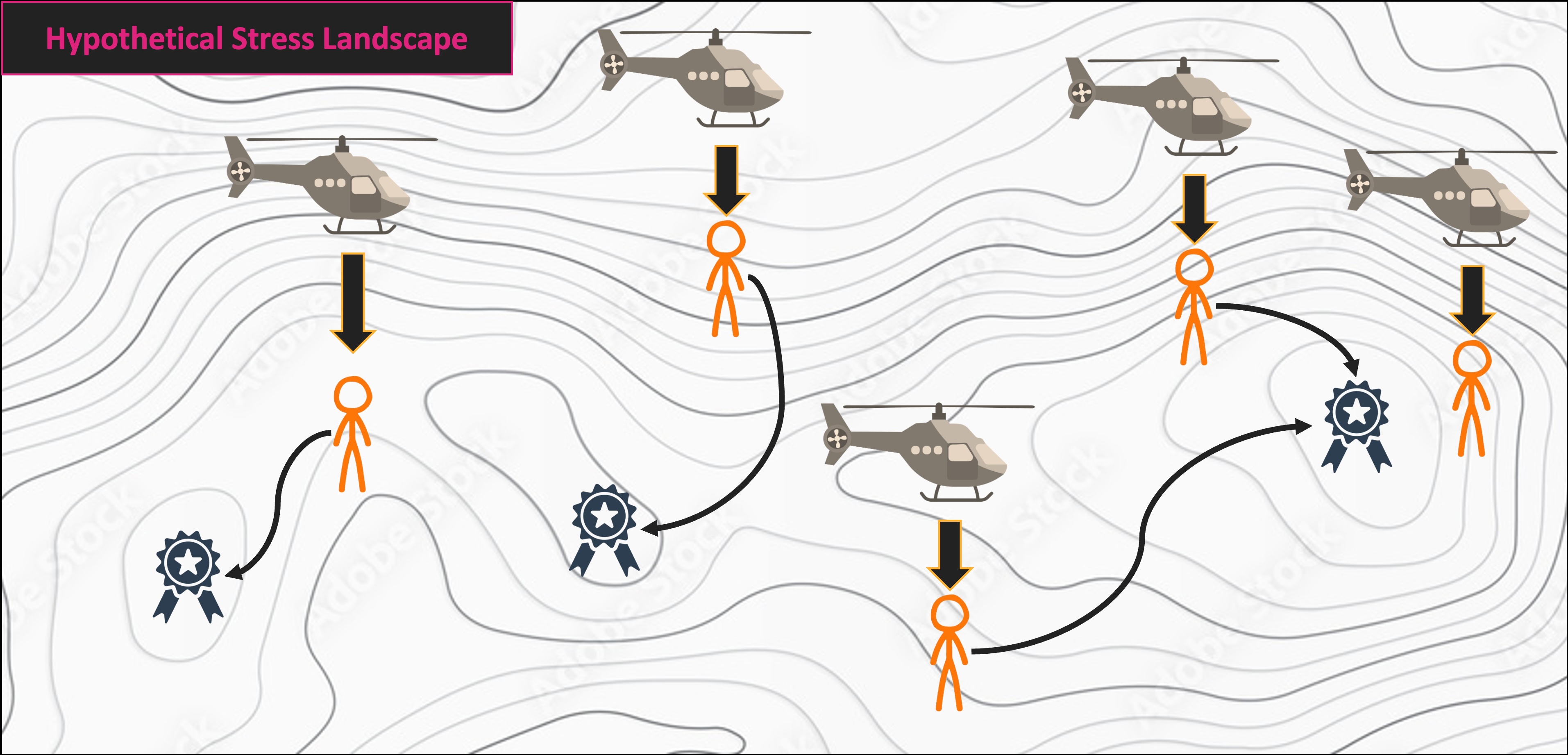
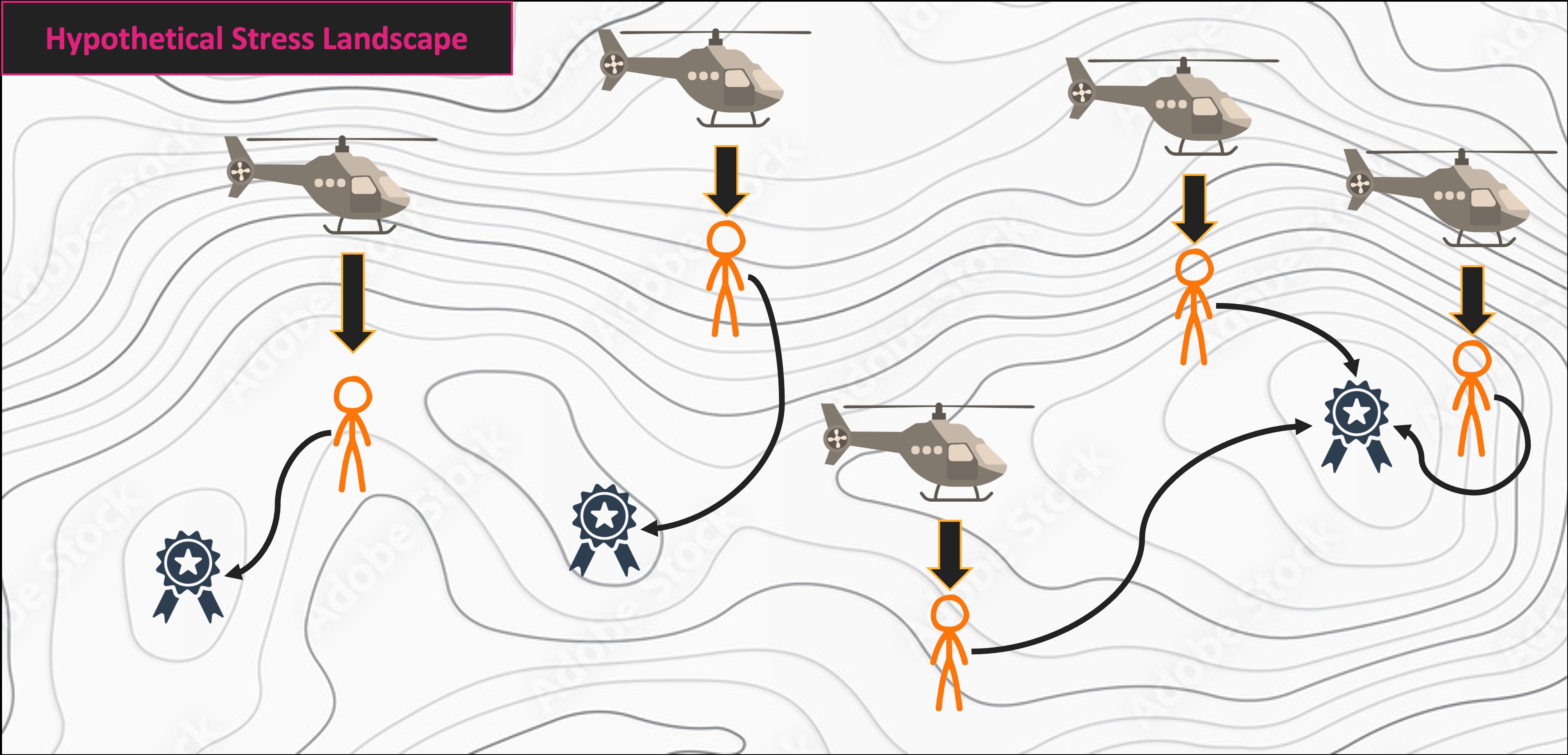
Multivariate Code Demo
Prepare
- First, you’ll need to install and load a few R packages
- While not technically necessary, the
librarianpackage makes library management much simpler
- While not technically necessary, the
Lichen Data
- The
veganpackage includes some lichen community composition data we can use for exploratory purposes
- We’ll begin by loading that and creating artificial groups for later analysis
# Load vegan's lichen dataset
utils::data("varespec", package = 'vegan')
# Make some columns of known number of groups
treatment <- c(rep.int("Trt1", (nrow(varespec)/4)),
rep.int("Trt2", (nrow(varespec)/4)),
rep.int("Trt3", (nrow(varespec)/4)),
rep.int("Trt4", (nrow(varespec)/4)))
# And combine them into a single data object
lichen_df <- cbind(treatment, varespec)Data Structure
- This data object now has the following structure:
'data.frame': 24 obs. of 45 variables:
$ treatment: chr "Trt1" "Trt1" "Trt1" "Trt1" ...
$ Callvulg : num 0.55 0.67 0.1 0 0 ...
$ Empenigr : num 11.13 0.17 1.55 15.13 12.68 ...
$ Rhodtome : num 0 0 0 2.42 0 0 1.55 0 0.35 0.07 ...
$ Vaccmyrt : num 0 0.35 0 5.92 0 ...
$ Vaccviti : num 17.8 12.1 13.5 16 23.7 ...
$ Pinusylv : num 0.07 0.12 0.25 0 0.03 0.12 0.1 0.1 0.05 0.12 ...
$ Descflex : num 0 0 0 3.7 0 0.02 0.78 0 0.4 0 ...
$ Betupube : num 0 0 0 0 0 0 0.02 0 0 0 ...
$ Vacculig : num 1.6 0 0 1.12 0 0 2 0 0.2 0 ...
$ Diphcomp : num 2.07 0 0 0 0 0 0 0 0 0.07 ...
$ Dicrsp : num 0 0.33 23.43 0 0 ...
$ Dicrfusc : num 1.62 10.92 0 3.63 3.42 ...
$ Dicrpoly : num 0 0.02 1.68 0 0.02 0.02 0 0.23 0.2 0 ...
$ Hylosple : num 0 0 0 6.7 0 0 0 0 9.97 0 ...
$ Pleuschr : num 4.67 37.75 32.92 58.07 19.42 ...
$ Polypili : num 0.02 0.02 0 0 0.02 0.02 0 0 0 0 ...
$ Polyjuni : num 0.13 0.23 0.23 0 2.12 1.58 0 0.02 0.08 0.02 ...
$ Polycomm : num 0 0 0 0.13 0 0.18 0 0 0 0 ...
$ Pohlnuta : num 0.13 0.03 0.32 0.02 0.17 0.07 0.1 0.13 0.07 0.03 ...
$ Ptilcili : num 0.12 0.02 0.03 0.08 1.8 0.27 0.03 0.1 0.03 0.25 ...
$ Barbhatc : num 0 0 0 0.08 0.02 0.02 0 0 0 0.07 ...
$ Cladarbu : num 21.73 12.05 3.58 1.42 9.08 ...
$ Cladrang : num 21.47 8.13 5.52 7.63 9.22 ...
$ Cladstel : num 3.5 0.18 0.07 2.55 0.05 ...
$ Cladunci : num 0.3 2.65 8.93 0.15 0.73 0.25 2.38 0.82 0.05 0.95 ...
$ Cladcocc : num 0.18 0.13 0 0 0.08 0.1 0.17 0.15 0.02 0.17 ...
$ Cladcorn : num 0.23 0.18 0.2 0.38 1.42 0.25 0.13 0.05 0.03 0.05 ...
$ Cladgrac : num 0.25 0.23 0.48 0.12 0.5 0.18 0.18 0.22 0.07 0.23 ...
$ Cladfimb : num 0.25 0.25 0 0.1 0.17 0.1 0.2 0.22 0.1 0.18 ...
$ Cladcris : num 0.23 1.23 0.07 0.03 1.78 0.12 0.2 0.17 0.02 0.57 ...
$ Cladchlo : num 0 0 0.1 0 0.05 0.05 0.02 0 0 0.02 ...
$ Cladbotr : num 0 0 0.02 0.02 0.05 0.02 0 0 0.02 0.07 ...
$ Cladamau : num 0.08 0 0 0 0 0 0 0 0 0 ...
$ Cladsp : num 0.02 0 0 0.02 0 0 0.02 0.02 0 0.07 ...
$ Cetreric : num 0.02 0.15 0.78 0 0 0 0.02 0.18 0 0.18 ...
$ Cetrisla : num 0 0.03 0.12 0 0 0 0 0.08 0.02 0.02 ...
$ Flavniva : num 0.12 0 0 0 0.02 0.02 0 0 0 0 ...
$ Nepharct : num 0.02 0 0 0 0 0 0 0 0 0 ...
$ Stersp : num 0.62 0.85 0.03 0 1.58 0.28 0 0.03 0.02 0.03 ...
$ Peltapht : num 0.02 0 0 0.07 0.33 0 0 0 0 0.02 ...
$ Icmaeric : num 0 0 0 0 0 0 0 0.07 0 0 ...
$ Cladcerv : num 0 0 0 0 0 0 0 0 0 0 ...
$ Claddefo : num 0.25 1 0.33 0.15 1.97 0.37 0.15 0.67 0.08 0.47 ...
$ Cladphyl : num 0 0 0 0 0 0 0 0 0 0 ...- Each column is an abbreviated lichen species name and the values are % cover
Permutation Analysis
- First, we’ll use the
RRPPpackage to use permutation analysis- HA: lichen community composition differs between at least two groups
- H0: lichen community composition does not differ
- Note that
RRPPdoes require a special class of data object to perform analysis
- Quick argument explanation:
- The
iterargument is the number of permutations - The
RRPPargument is whether to permute residuals (TRUE) or the full data (FALSE)
- The
Permutation Analysis Results
- To check the results, we can use the
anovafunction- This also allows us to specify the desired effect type
# Check out the results of the analysis!
RRPP::anova.lm.rrpp(lich_fit, effect.type = "F", print.progress = F)
Analysis of Variance, using Residual Randomization
Permutation procedure: Randomization of null model residuals
Number of permutations: 1000
Estimation method: Ordinary Least Squares
Sums of Squares and Cross-products: Type I
Effect sizes (Z) based on F distributions
Df SS MS Rsq F Z Pr(>F)
treatment 3 19602 6533.9 0.46682 5.8369 3.655 0.001 **
Residuals 20 22388 1119.4 0.53318
Total 23 41990
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Call: RRPP::lm.rrpp(f1 = community ~ treatment, iter = 999, RRPP = T,
data = lichen_rpdf)- Now we get a fairly standard ANOVA table
- The
Zcolumn is the “Z score” and is essentially the effect size
- The
Permutation Pairwise Comparisons
- Once we have our main results, we can also get pairwise comparison results
# Perform pairwise comparisons
lich_pairs <- RRPP::pairwise(fit = lich_fit, groups = treatment)
# Check results
summary(lich_pairs)
Pairwise comparisons
Groups: Trt1 Trt2 Trt3 Trt4
RRPP: 1000 permutations
LS means:
Vectors hidden (use show.vectors = TRUE to view)
Pairwise distances between means, plus statistics
d UCL (95%) Z Pr > d
Trt1:Trt2 16.48128 39.64863 -0.7289177 0.760
Trt1:Trt3 38.04299 40.10757 1.5478150 0.067
Trt1:Trt4 59.41369 38.99550 2.9255939 0.001
Trt2:Trt3 37.15261 38.01919 1.5148939 0.063
Trt2:Trt4 62.40360 39.28595 3.0093479 0.001
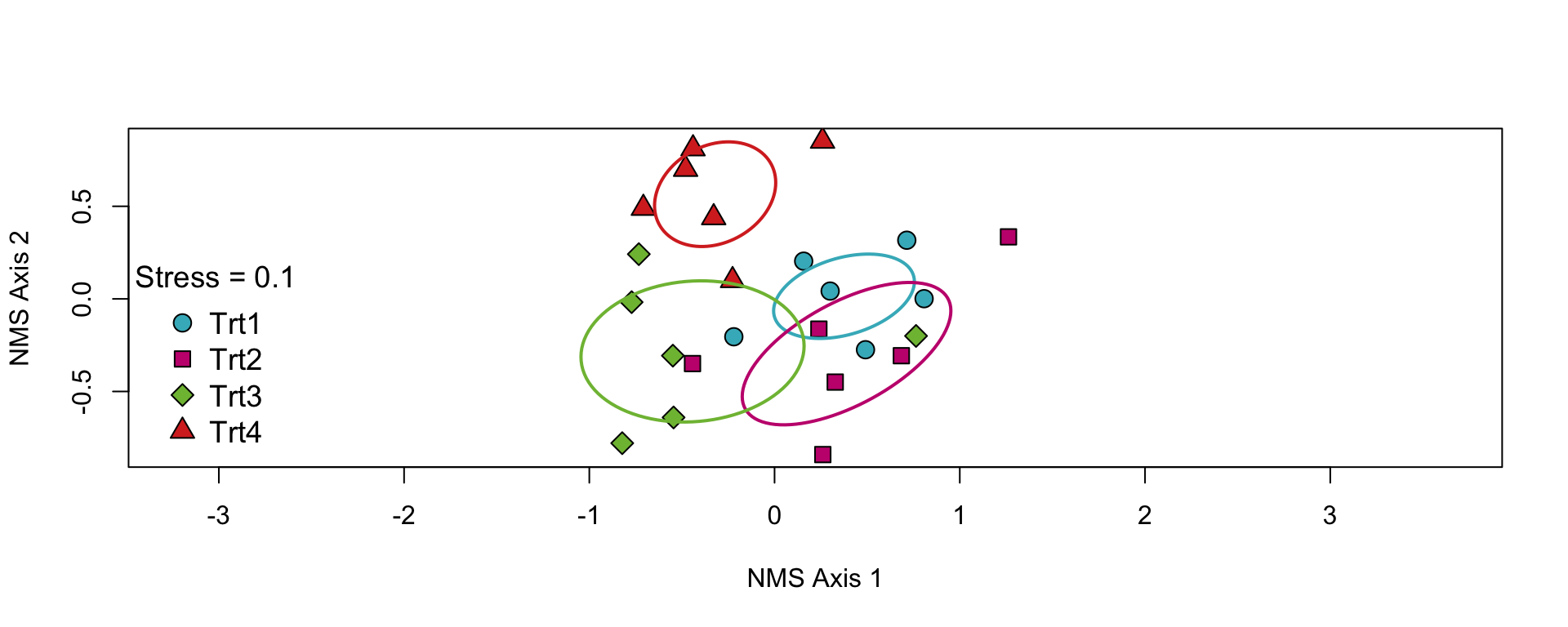
Trt3:Trt4 50.44284 40.37863 2.2553908 0.005- These show that the fourth treatment significantly differs from the other three
- And the third marginally differs from the first and second
Multivariate Data Visualization
- There are a few non-ordination ways of doing multivariate visualization
- Note that these are mostly for exploratory purposes
- They’re still helpful for checking the general ‘vibe’ of the data
- but they may not hold up to formal review processes
3D Scatterplot
- Perhaps the simplest mode of multivariate data visualization is just to make a 3D scatterplot!
- Still technically counts as “multivariate” visualization
- Primary benefit is that interpretation is pretty straightforward
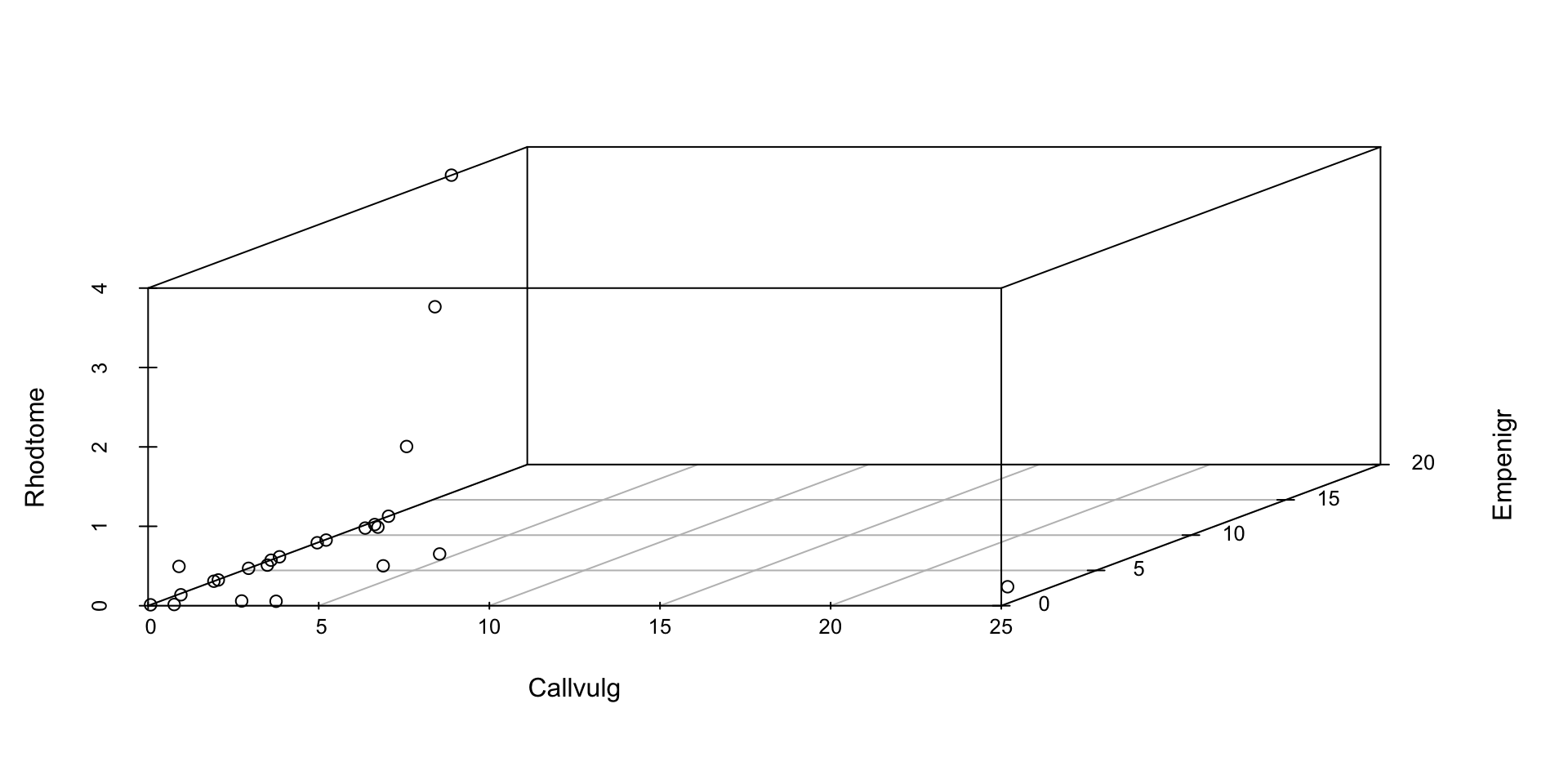
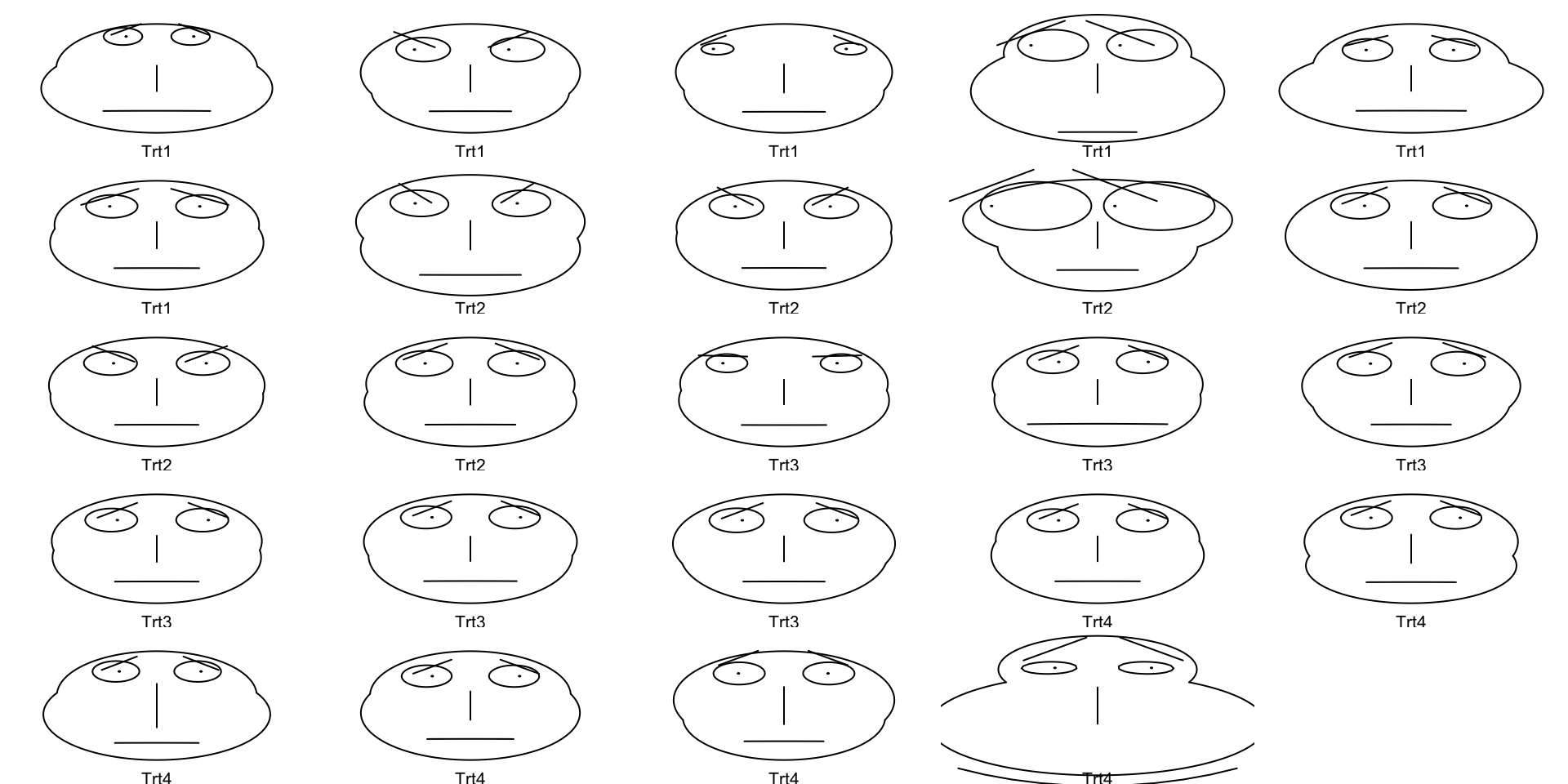
Chernoff Faces
- Some people have tried to make human’s capacity for comparing faces into a tool for data visualization
- Data are transformed into human(-ish) faces with different dimensions
- I find these very scary
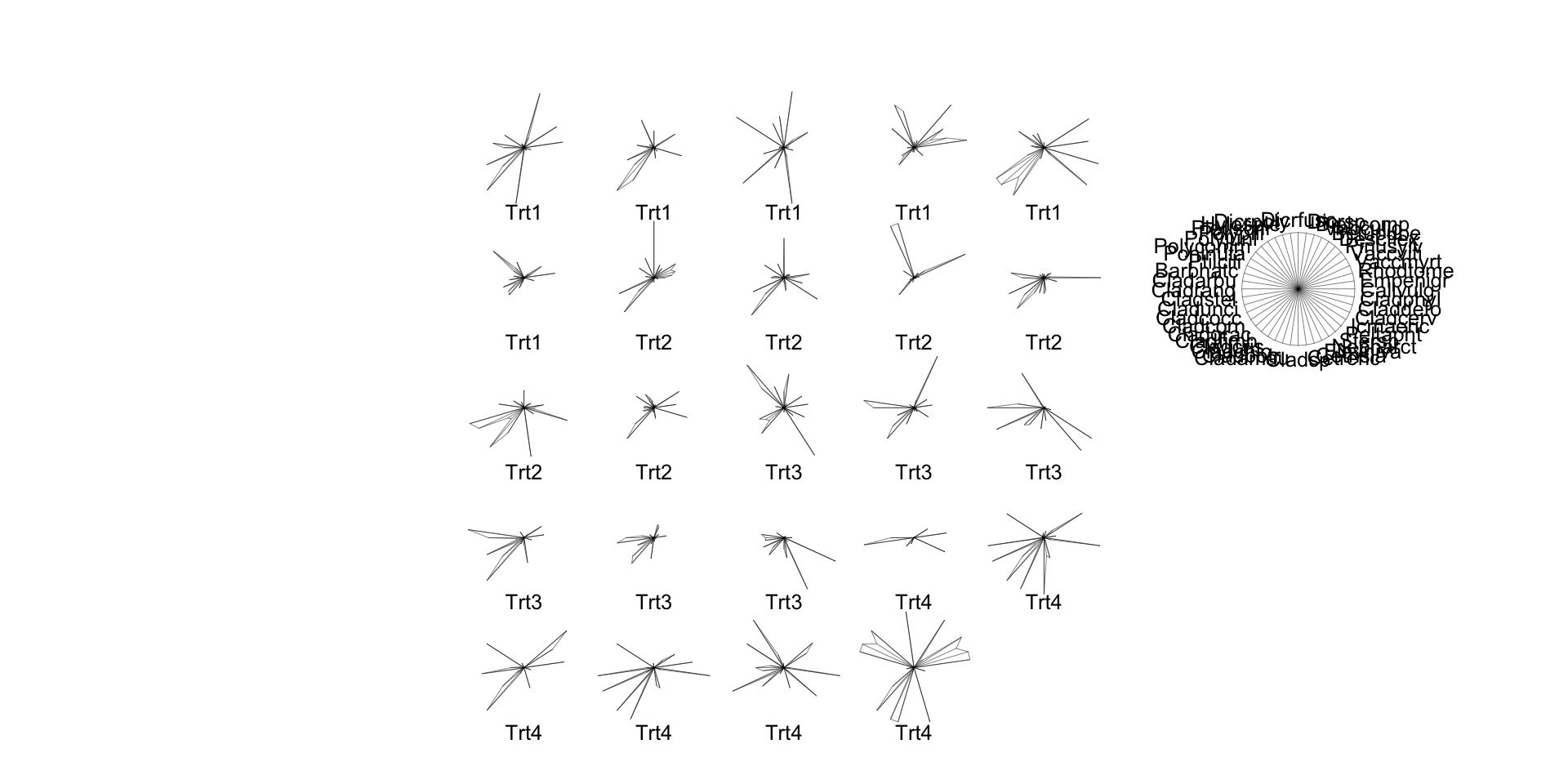
Star Plots
- Perhaps most usefully, you can just make “star plots” to check multivariate data
Principal Components
- Before we can visualize PCA results, we need to actually identify PC axes!
# Perform Principal Components Analysis
lich_pc <- prcomp(x = varespec)
# Summarize it to calculate '% variation explained' for each PC axis
lich_pc_smry <- summary(lich_pc)
# Check the structure of the summarized object
str(lich_pc_smry)List of 6
$ sdev : num [1:24] 31.35 21.55 11.5 8.6 6.96 ...
$ rotation : num [1:44, 1:24] -0.01399 0.01566 -0.00646 -0.05168 0.00858 ...
..- attr(*, "dimnames")=List of 2
.. ..$ : chr [1:44] "Callvulg" "Empenigr" "Rhodtome" "Vaccmyrt" ...
.. ..$ : chr [1:24] "PC1" "PC2" "PC3" "PC4" ...
$ center : Named num [1:44] 1.88 6.33 0.35 2.11 11.46 ...
..- attr(*, "names")= chr [1:44] "Callvulg" "Empenigr" "Rhodtome" "Vaccmyrt" ...
$ scale : logi FALSE
$ x : num [1:24, 1:24] -10.8 -27.8 -25.7 -31.8 -19.6 ...
..- attr(*, "dimnames")=List of 2
.. ..$ : chr [1:24] "18" "15" "24" "27" ...
.. ..$ : chr [1:24] "PC1" "PC2" "PC3" "PC4" ...
$ importance: num [1:3, 1:24] 31.352 0.538 0.538 21.548 0.254 ...
..- attr(*, "dimnames")=List of 2
.. ..$ : chr [1:3] "Standard deviation" "Proportion of Variance" "Cumulative Proportion"
.. ..$ : chr [1:24] "PC1" "PC2" "PC3" "PC4" ...
- attr(*, "class")= chr "summary.prcomp"Principal Components Ordination
# With that done, we can make a graph of that information!
plot(x = lich_pc$x[,1], y = lich_pc$x[,2], pch = 20,
## And do some fancy axis labels to get 'variation explained' in the plot
xlab = paste0("PC1 (", (lich_pc_smry$importance[2, 1] * 100), " % variation explained)"),
ylab = paste0("PC2 (", (lich_pc_smry$importance[2, 2] * 100), " % variation explained)"))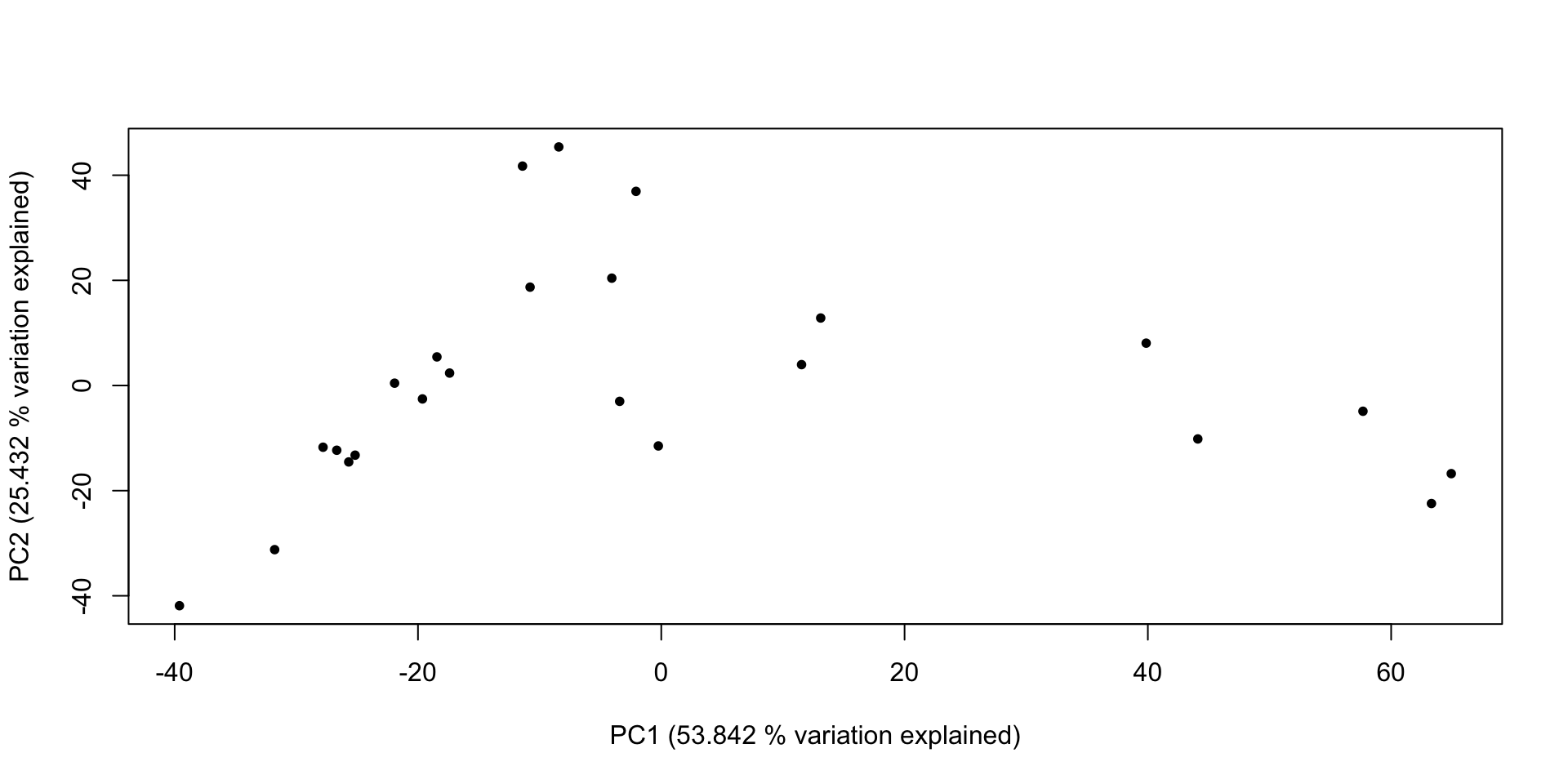
Non-Metric Multidimensional Scaling
- Just like PCA, we need to start by actually performing the scaling step
- See here:
- Quick explanation of (some of) the arguments
distanceis your preferred metric for dissimilaritykis the number of axes to scale to (typically 2 for standard plotting)tryis the number of starting data configurations (remember the helicopter analogy!)
NMS Ordination
- Fortunately, we can use a nice ordination function from the
supportRpackage to make the graph